Place the Steps in the Correct Order to Review the Steps of Mrna Processing in Eukaryotes
five.5: RNA Processing
- Page ID
- 1654
So far, we accept looked at the machinery past which the information in genes (DNA) is transcribed into RNA. The newly made RNA, likewise known as the primary transcript (the product of transcription is known as a transcript) is further processed before information technology is functional. Both prokaryotes and eukaryotes procedure their ribosomal and transfer RNAs.
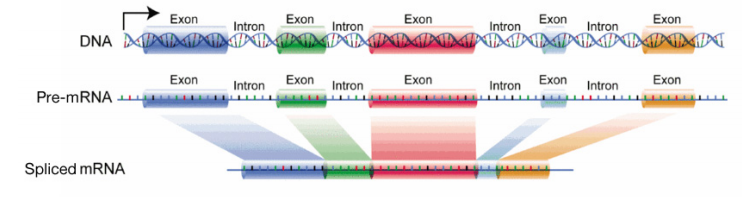
The major divergence in RNA processing, however, between prokaryotes and eukaryotes, is in the processing of messenger RNAs. We will focus on the processing of mRNAs in this word. You lot volition recall that in bacterial cells, the mRNA is translated straight equally it comes off the DNA template. In eukaryotic cells, RNA synthesis, which occurs in the nucleus, is separated from the poly peptide synthesis machinery, which is in the cytoplasm. In addition, eukaryotic genes have introns, noncoding regions that interrupt the factor'south coding sequence. The mRNA copied from genes containing introns will also therefore accept regions that interrupt the information in the gene. These regions must be removed before the mRNA is sent out of the nucleus to be used to direct protein synthesis. The process of removing the introns and rejoining the coding sections or exons, of the mRNA, is chosen splicing. In one case the mRNA has been capped, spliced and had a polyA tail added, information technology is sent from the nucleus into the cytoplasm for translation.
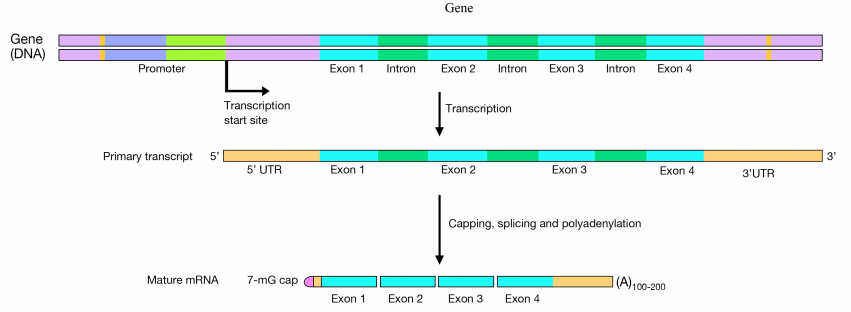
The initial product of transcription of a protein coding gene is called the pre-mRNA (or primary transcript). After it has been candy and is gear up to be exported from the nucleus, it is called the mature mRNA or processed mRNA.
What are the processing steps for messenger RNAs?
In eukaryotic cells, pre-mRNAs undergo three main processing steps:
- Capping at the 5' end
- Addition of a polyA tail at the 3' finish. and
- Splicing to remove introns
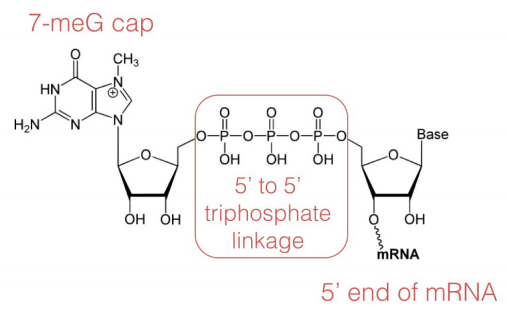
In the capping step of mRNA processing, a 7-methyl guanosine (shown at left) is added at the 5' terminate of the mRNA. The cap protects the v' end of the mRNA from deposition by nucleases and also helps to position the mRNA correctly on the ribosomes during protein synthesis.
The 3' terminate of a eukaryotic mRNA is first trimmed, then an enzyme called PolyA Polymerase adds a "tail" of about 200 'A' nucleotides to the 3' end. There is evidence that the polyA tail plays a role in efficient translation of the mRNA, as well every bit in the stability of the mRNA. The cap and the polyA tail on an mRNA are as well indications that the mRNA is consummate (i.east., not defective). Introns are removed from the pre-mRNA by the activity of a circuitous called the spliceosome. The spliceosome is fabricated up of proteins and modest RNAs that are associated to form protein-RNA enzymes called small-scale nuclear ribonucleoproteins or snRNPs (pronounced SNURPS). The splicing machinery must be able to recognize splice junctions (i.e., the terminate of each exon and the showtime of the next) in club to correctly cut out the introns and join the exons to make the mature, spliced mRNA.
What signals indicate where an intron starts and ends? The base sequence at the kickoff (v' or left stop, also chosen the donor site) of an intron is GU while the sequence at the three' or right finish (a.g.a. acceptor site) is AG. In that location is likewise a 3rd important sequence within the intron, chosen a co-operative point, that is of import for splicing.
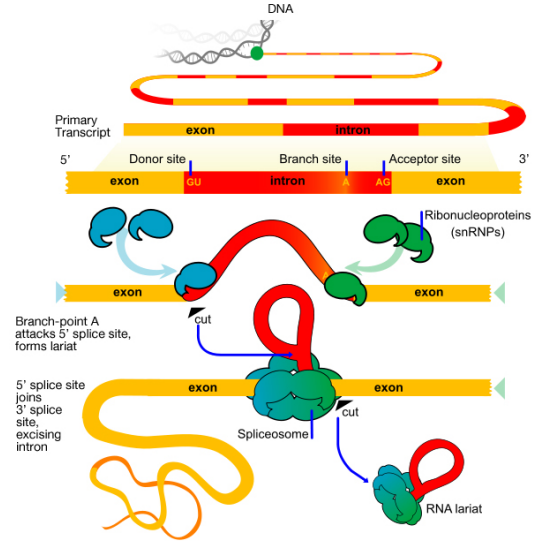
At that place are two main steps in splicing:
- In the beginning step, the pre-mRNA is cutting at the 5' splice site (the junction of the 5' exon and the intron). The 5' end of the intron and then is joined to the co-operative point within the intron. This generates the lariat-shaped molecule characteristic of the splicing process
- In the second step, the 3' splice site is cut, and the two exons are joined together, and the intron is released.
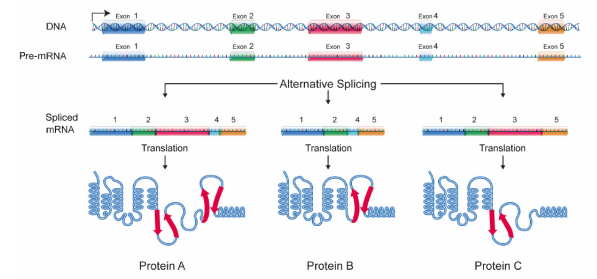
Many pre-mRNAs have a large number of exons that can be spliced together in unlike combinations to generate different mature mRNAs. This is called alternative splicing, and allows the production of many different proteins using relatively few genes, since a single RNA tin, by combining unlike exons during splicing, create many different protein coding messages. Considering of culling splicing, each gene in our Dna gives rise, on average, to three different proteins. Once protein coding messages have been processed by capping, splicing and improver of a poly A tail, they are transported out of the nucleus to exist translated in the cytoplasm.
Source: https://bio.libretexts.org/Bookshelves/Biochemistry/Book%3A_Biochemistry_Free_and_Easy_(Ahern_and_Rajagopal)/05%3A_Flow_of_Genetic_Information/5.05%3A_RNA_Processing
0 Response to "Place the Steps in the Correct Order to Review the Steps of Mrna Processing in Eukaryotes"
Post a Comment